Pytorch implementation of the paper Bayesian Generative Models for Knowledge Transfer in MRI Semantic Segmentation Problems
We use Deep Weight Prior (DWP) [1] to perform transfer learning from large source to the smaller target dataset with medical images.
We use a common benchmark - BRATS18 [2] as a target and MS [3] dataset as source dataset.
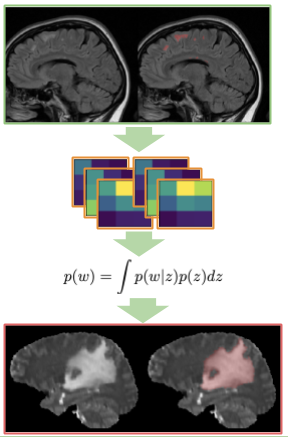
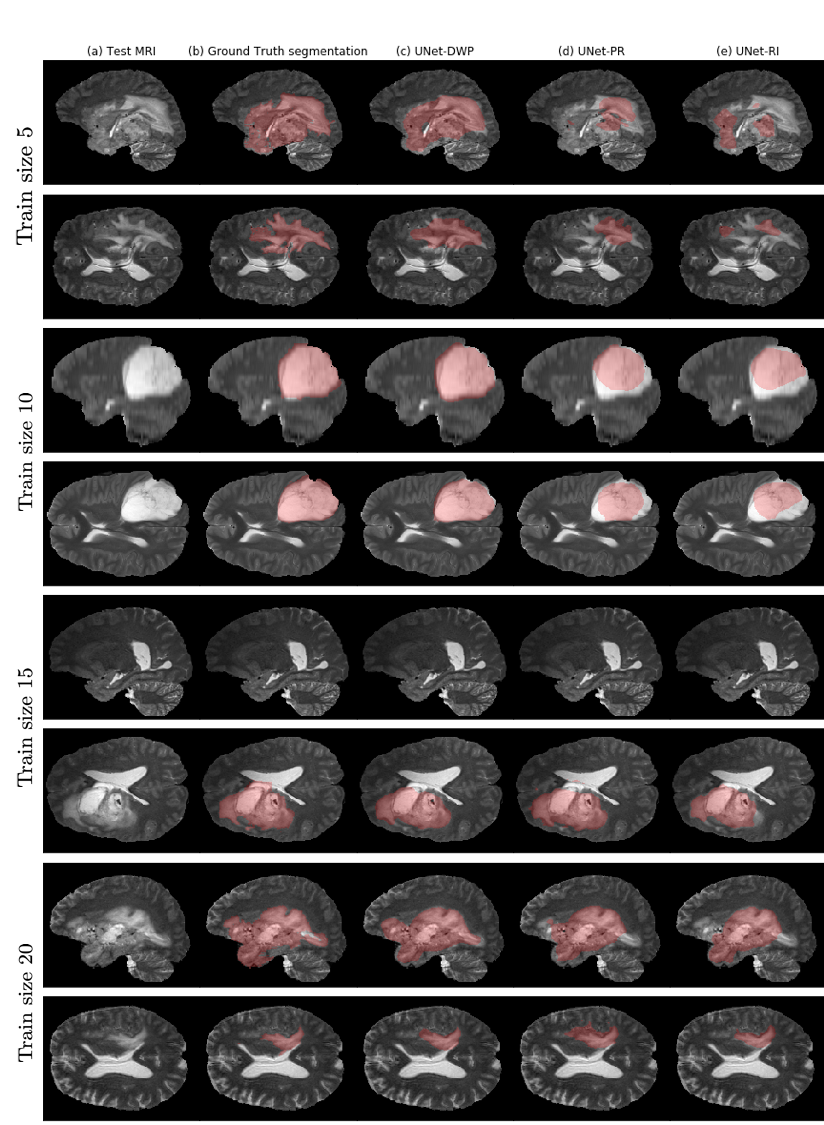
The method perform better that randomly initialized and fine-tuned models.
[1] Atanov, A., Ashukha, A., Struminsky, K., Vetrov, D., and Welling, M. (2018). The deep weight prior.
[2] Menze, B. H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., et al. (2015). The multimodal brain tumor image segmentation benchmark (brats).
[3] CoBrain analytics, (2018). Multiple Sclerosis Human Brain MR Imaging Dataset
Experiments
Environment setup
The code is base on PyTorch 1.5.0 and PyTorch-Lightning 0.8.0.
The exact specification of our environment is provided in the file environment.yml and
can be created via
conda env create -f environment.yml
Example on toy dataset
-
Load Source and target dataset into
data/dataset_namefolders. Below are examples forMNISTabdnotMNISTdatasets (they will be loaded automatically) - Train N models on the source dataset. Here full notMNIST dataset is used
python3 train.py --dataset_name notMNIST --trian_size -1 - Train VAE on the kernels
python3 train_vae.py --kernel_size 7 - Train model on the target dataset with VAE from the previous step as a prior
python3 train.py --dataset_name MNIST --prior notMNIST --trian_size 100
Citation
If you find our paper or code useful, feel free to cite the paper:
@article{kuzina2019bayesian,
title={Bayesian generative models for knowledge transfer in mri semantic segmentation problems},
author={Kuzina, Anna and Egorov, Evgenii and Burnaev, Evgeny},
journal={Frontiers in neuroscience},
volume={13},
pages={844},
year={2019},
publisher={Frontiers}
}